Run DICAST with a graphical user interface (depreciated)
Make sure you are in the working directory and that listing your directory looks like the directory structure mentioned in directory structure. To run dicast, activate the dicast-snakemake conda environment:
conda activate dicast-snakemake
Your prompt should show you (dicast-snakemake), to show you your conda environment. If so, start DICAST with the following command.
python gui/dicast.py
Warning
DICAST is set to Run ASimulatoR with default values, however, should you wish to tune simulated dataset to your investigative questions, please modify the file scripts/ASimulatoR_config.R ( See ASimulatoR Parameters )
Note
The graphical user interface assumes a X11 rendering system. If you’re using ssh, please use the ssh -X flag, to allow X11 forwarding. If you’re not on a Linux machine, locally, find out how to host an X11 daemon for yourself. If you’re on a Mac, this could mean installing Xquartz on your localmachine. If you’re running DICAST on your local linux machine, the output of echo $DISPLAY should read :0; this suggests that you have X11 forwarded correctly to your localmachine.
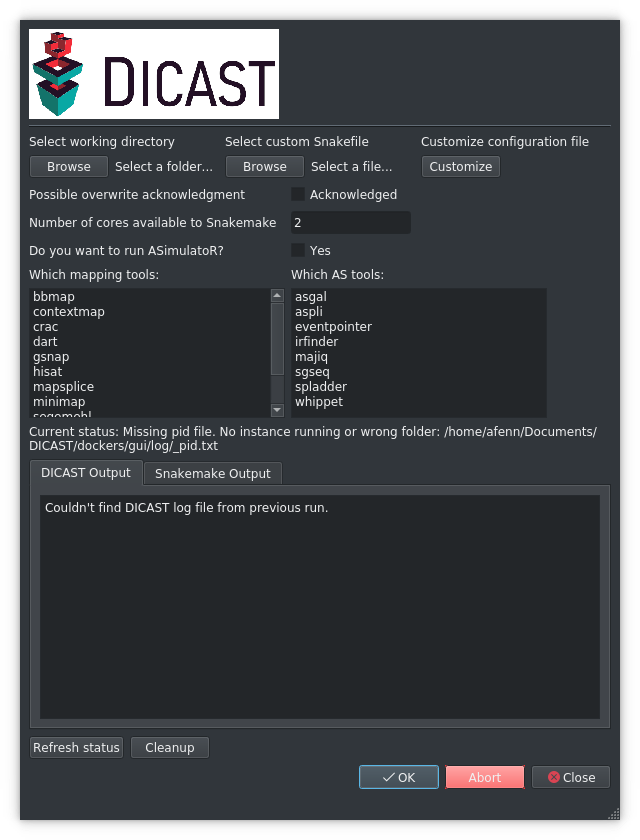
Options |
Explanation |
|---|---|
Select working directory |
The working directory is where you’ve hosted your DICAST git and it looks like the described directory structure needed to run DICAST. This acts as the root directory for the project. |
Select custom Snakefile |
By default, DICAST stores it’s Snakefile under |
Possible overwrite acknowledgement |
DICAST writes all outputs to |
Number of cores available to Snakemake |
Total number of cores given to DICAST. Minimum advised: 2 |
Do you want to run ASimulatoR? |
ASimulatoR comes as a part of DICAST. ASimulatoR can be configured by modifying file at |
Referesh status |
Is useful, if you connect to DICAST and a previous run is still continuing. DICAST catches previous run and updates your progress. |
Abort |
Interrupt a running DICAST execution. |
Clean up |
Cleans up intermediate snakemake files from incomplete runs, use after Abort. |
Which Mapping/AS tools: |
Select the tools you want to run within DICAST and click okay. |
OK |
Starts a DICAST run. |
Close |
Shuts down the GUI, leaving a running session of DICAST, you can connect to next time you start the gui. |
DICAST will continue to run, and you can safely close the GUI. Re-opening the GUI connects you back to a running instance of DICAST, if it isn’t finished already.
Custom Configuration file:
With this, custom configure your dicast run

Options |
Explanation |
|---|---|
Number of cored for each tools |
Usually seen as –ncores for many tools. gives the option of efficient parallelization when possible. |
Read length |
An option seen by quite a few tools. supported length <200. Not every tool has been tested with varying read lengths. |
Fasta name |
Browse to select your reference genome |
GTF name |
Browse to select your reference annotation, if you’re using a real dataset. If you’re using ASimulatoR, leave this as |
GFF name |
Browse to select your reference annotation, if you’re using a real dataset. If you’re using ASimulatoR, leave this as |
First fastq pair suffix |
How do you differentiate paired end reads? how does each fastq file end? “1.fastq”? |
Second fastq pair suffix |
How do you differentiate paired end reads? how does each fastq file end? “2.fastq”? |
Base output directory |
Please leave this at default |
Use BAM files as inputs |
Some tools give you the option of starting from fastq files or from mapped files. |
Should different AS events be combined into one |
Multiple exon skipping, could be considered one or many AS events. How do you prefer reporting them? |
Note
the close button warns you about losing changes even if you saved them. This is a bug, and will be corrected soon.
Troubleshooting
Check Snakemake Output to see which rule failed.
if the rule that failed was named after a tool, check log files under output/<tool>-output/logs/ to see where the error was.
Warning
Aborting a run: Once the dockers begin, they’re not under DICAST’s control to abort. If you really want to interrupt DICAST, also check for running containers docker ps and stop/ kill running containers with docker stop <container-name>. Also use the clean up function to clean up an interrupted run.
Interrupting a DICAST run
If you want to interrupt a DICAST run. Click on the Abort button and then click on the Clean up button. DICAST unfortunately doesn’t show you that this is a required step, so until Clean up is clicked upon, your next run will not start. Your configurations should stay as you set them last. Click on Acknowledge overwrite checkbox and you’re all set for the next run.