Welcome to DICAST!
The DICAST pipeline was initially designed to benchmark alternative splicing (AS) tools based on simulated “ground truth” reads produced by ASimulator. Here we provide a pipeline for running several mapping and AS event detection tools and evaluate and compare the results. DICAST however is not only suitable for simulated data, but you can also use it for real data.
We hope for this to be an open collaborative effort to represent and benchmark your tools. Please feel free to reach out to us, should your tool already be here and you have some edits to suggest. Also please reach out to us, if your Alternative splicing tool isn’t here and you’d like it to be.
To provide a fair baseline while maintaining easy usability, per default we run the tools with their default variables. If you feel like this is not doing your tool justice please contact us. The default parameters can be changed by editing the ENTRYPOINT.sh scripts of each tool.
The tools included here are the most widely used and well maintained among AS event detection tools. If you would like to include your tool in the pipeline please let us know. We hope this collection can be a starting point for future benchmarking approaches and quality control.
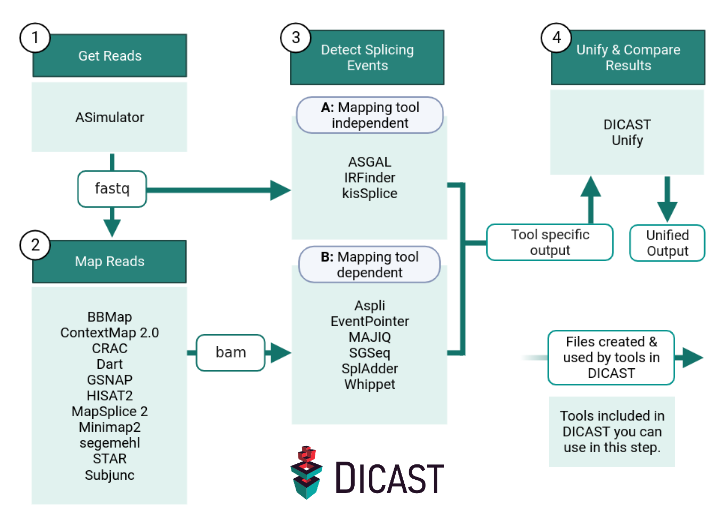
What is DICAST?
DICAST is a collection of alternative splicing event detection tools for analyzing RNA-Seq data. DICAST runs on Snakemake pipelines and relies on Docker based containerization. For easy installation and maintenance, we provide docker containers for every integrated tool at Dockerhub
DICAST can be run as a complete pipeline, starting with simulating RNA-Seq data with ASimulator, mapping the reads to a fasta reference, get information about AS with one or multiple tools and finally visualize and compare the results from different tools with DICAST unify.
Alternatively, you can run one of the same tools as a single docker container without snakemake.
DICAST is available for download on Github.
When should I use DICAST?
If you want to benchmark or compare different mapping and splicing tools with a genome and an annotation.
If you want to analyze your data with one or more tools included in DICAST and find out which tools give you the events you’re interested in.
How do I cite DICAST?
The preprint citing DICAST is Alternative splicing analysis benchmark with DICAST, is available for review on bioRxiv
- If you use DICAST please cite the preprint as:
|Fenn, A.M., Tsoy, O., Faro, T., Roessler, F., Dietrich, A., Kersting, J., Louadi, Z., Lio, C.T., Voelker, U., Baumbach, J. and Kacprowski, T., 2022. Alternative splicing analysis benchmark with DICAST. bioRxiv.